Welcome to CATMAID¶
The Collaborative Annotation Toolkit for Massive Amounts of Image Data
Features:
Fast terabyte-scale image data browsing
Collaborative microcircuit reconstruction and annotation
Flexible hierarchical semantic annotation
Multiple linked image stack display
Neuron Catalog
SVG and WebGL-based neuronal morphology viewer
Open source software (GPLv3)
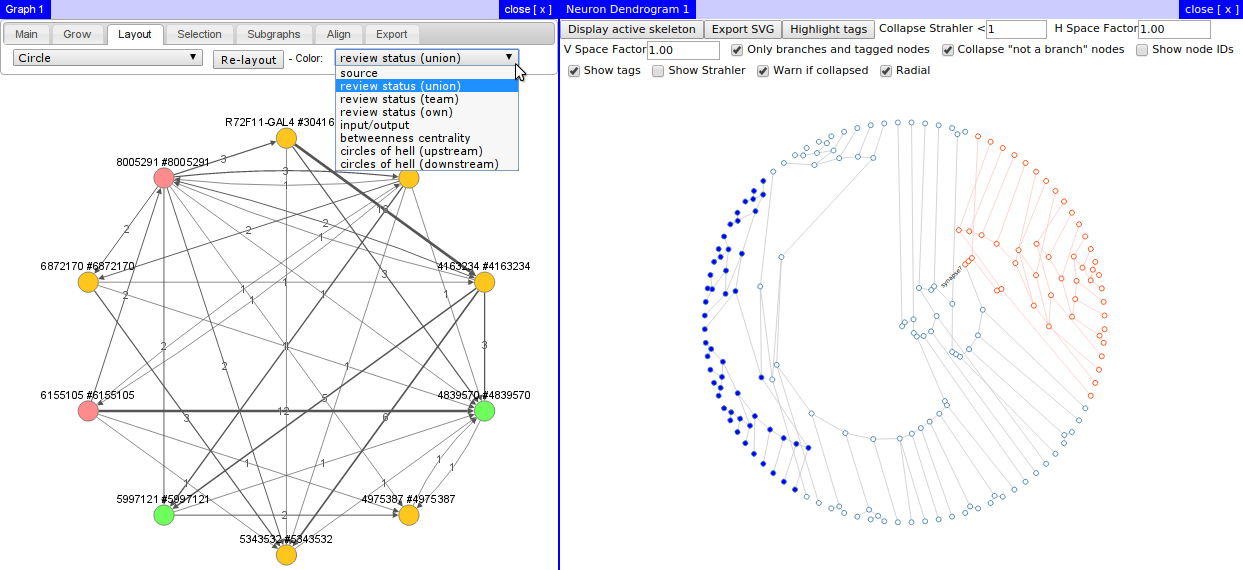
User interface
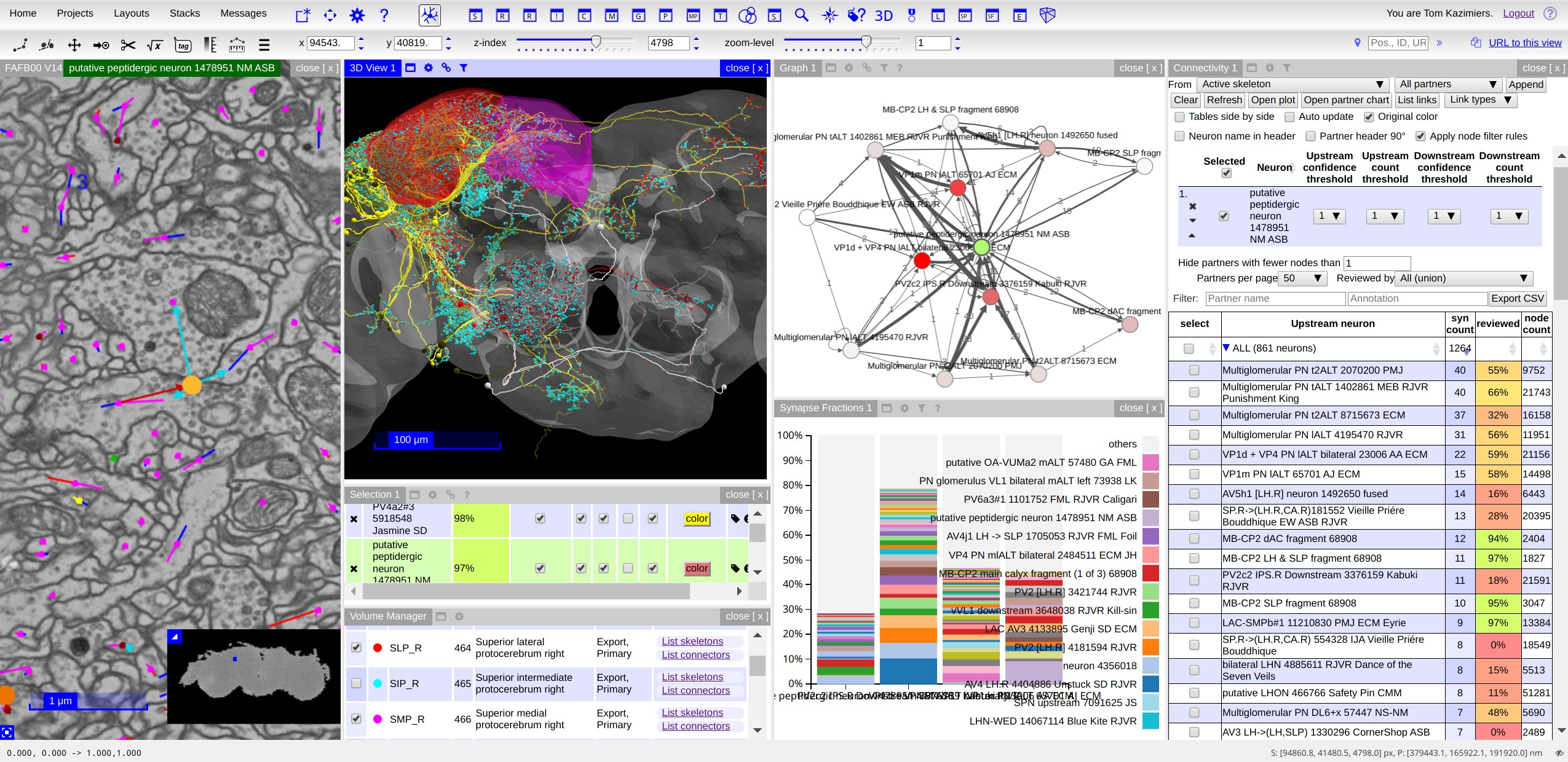
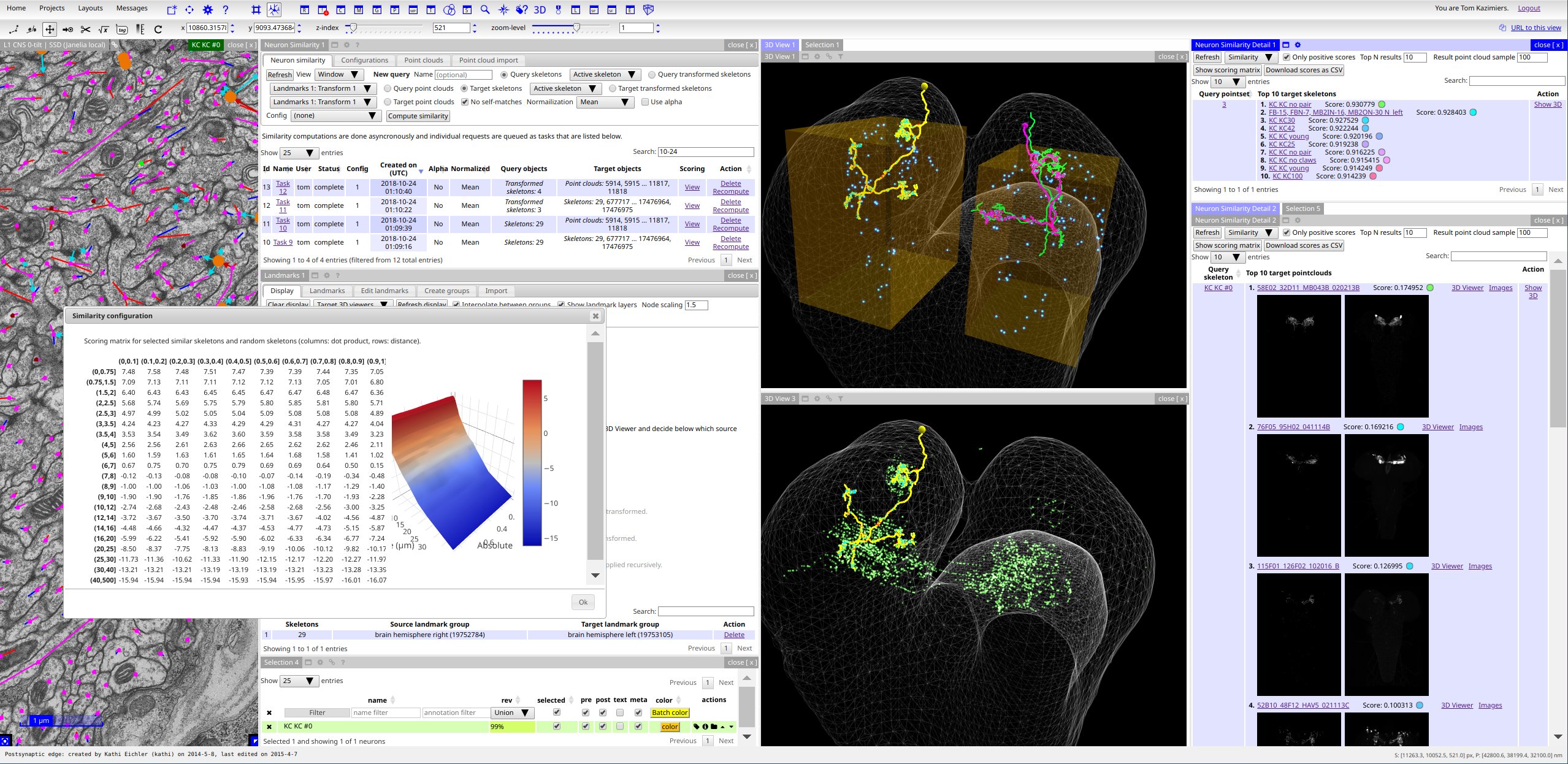


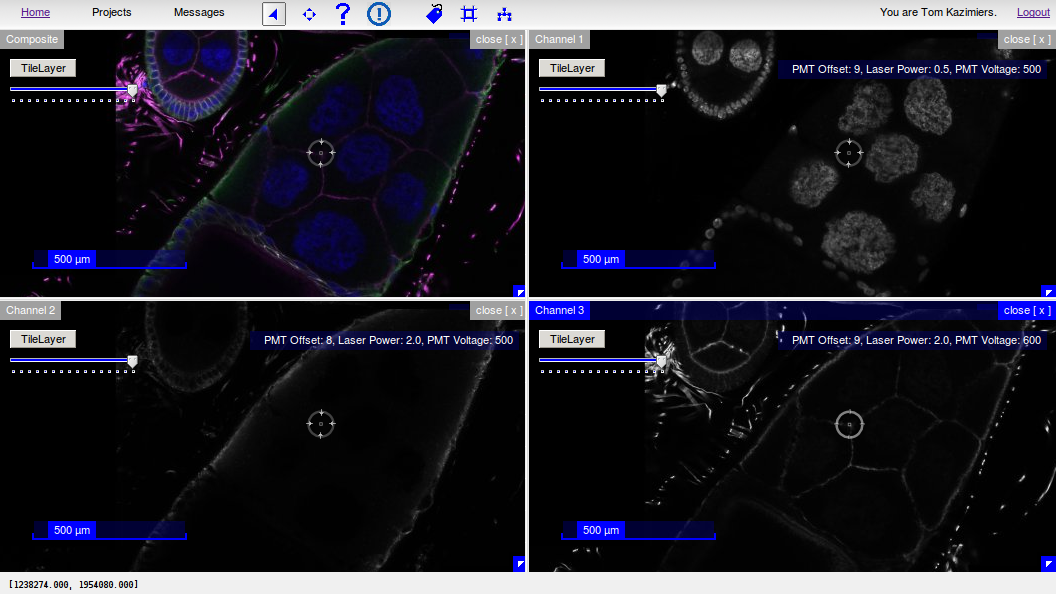

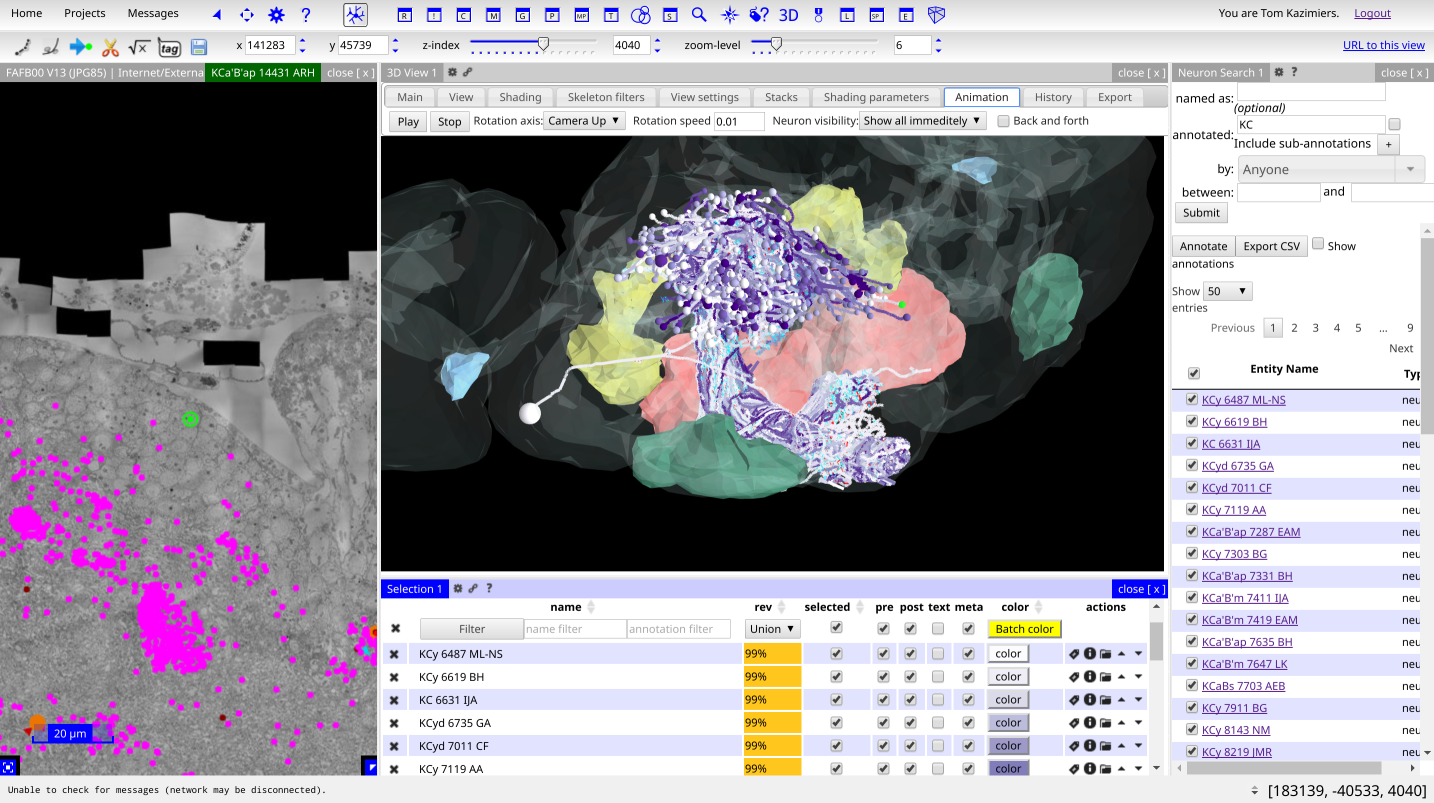

Documentation¶
- User Documentation
- Administrator Documentation
- Basic Installation Instructions
- Configuration options
- Administering a CATMAID Instance
- Setting Up Nginx for CATMAID
- Example configurations
- Database backups
- Node providers
- Permissions and access control
- Installation of the Celery task queue
- WebSockets and ASGI
- Tracing data caching
- History and provenance tracking
- Data replication
- Postgres administration
- Docker
- User accounts
- Additional back-ends
- R setup for NBLAST and related tools
- Configuring NBLAST
- External sign-in using OAuth2
- Frequently Asked Questions
- Installing Catmaid with an RDS database backend
- Developer Documentation
- Developer setup with Vagrant
- Contributing to CATMAID
- Tile Source Conventions
- Creating widgets
- Creating New Data View Types
- Deep Link URL Format
- Database Migrations
- Django Unit Tests for CATMAID
- Models, State and Commands
- Creating a CATMAID Instance on EC2
- Creating CATMAID extensions
- CATMAID Releases
