Screenshots and videos¶
Tom Kazimiers with an introduction to CATMAID at NEUBIAS Academy 2021
Stephan Saalfeld talking about CATMAID, TrakEM2 and ImgLib2
Demonstration of some of the neuron shading options in CATMAID's 3D viewer (v2015.12.12)
This is the central settings widget of CATMAID (v2015.12.12).
Clustering widget for ontology based classification graphs (v2015.12.12)
3D view of the 72 hpf HT9-4 larva showing fully-traced peptidergic neurons identified by siGOLD. This movie was published under a CC-BY Licence and was created by Shaidi et. al. as part of the publication "A serial multiplex immunogold labeling method for identifying peptidergic neurons in connectomes", published 15 Dec 2015 in eLife.
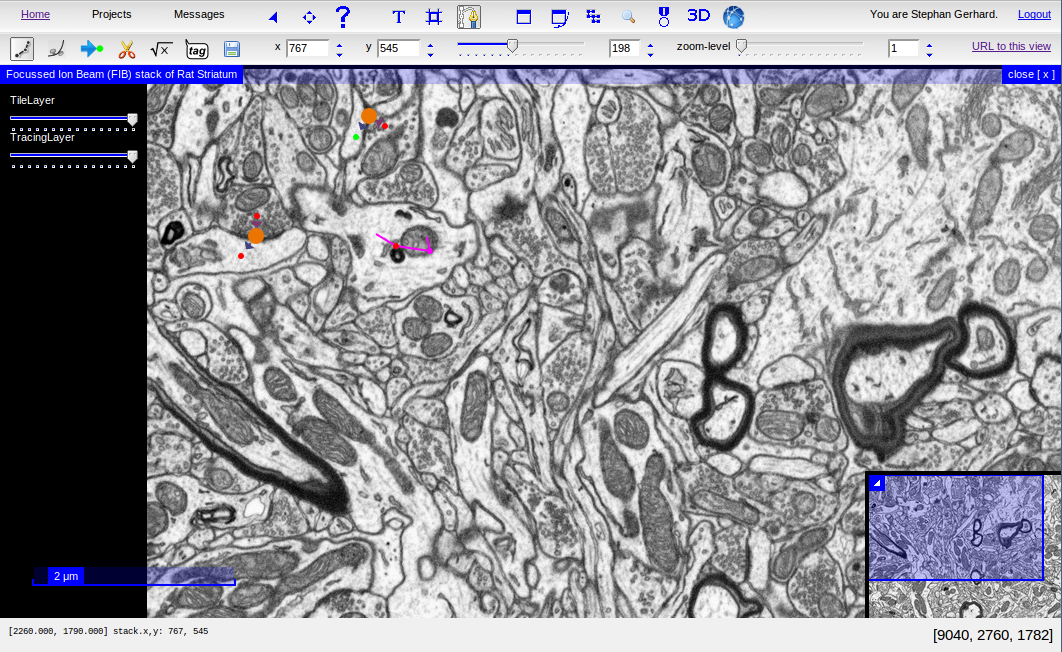
The CATMAID User Interface with skeleton annotations and tags¶

CATMAID comes with a 3D viewer¶
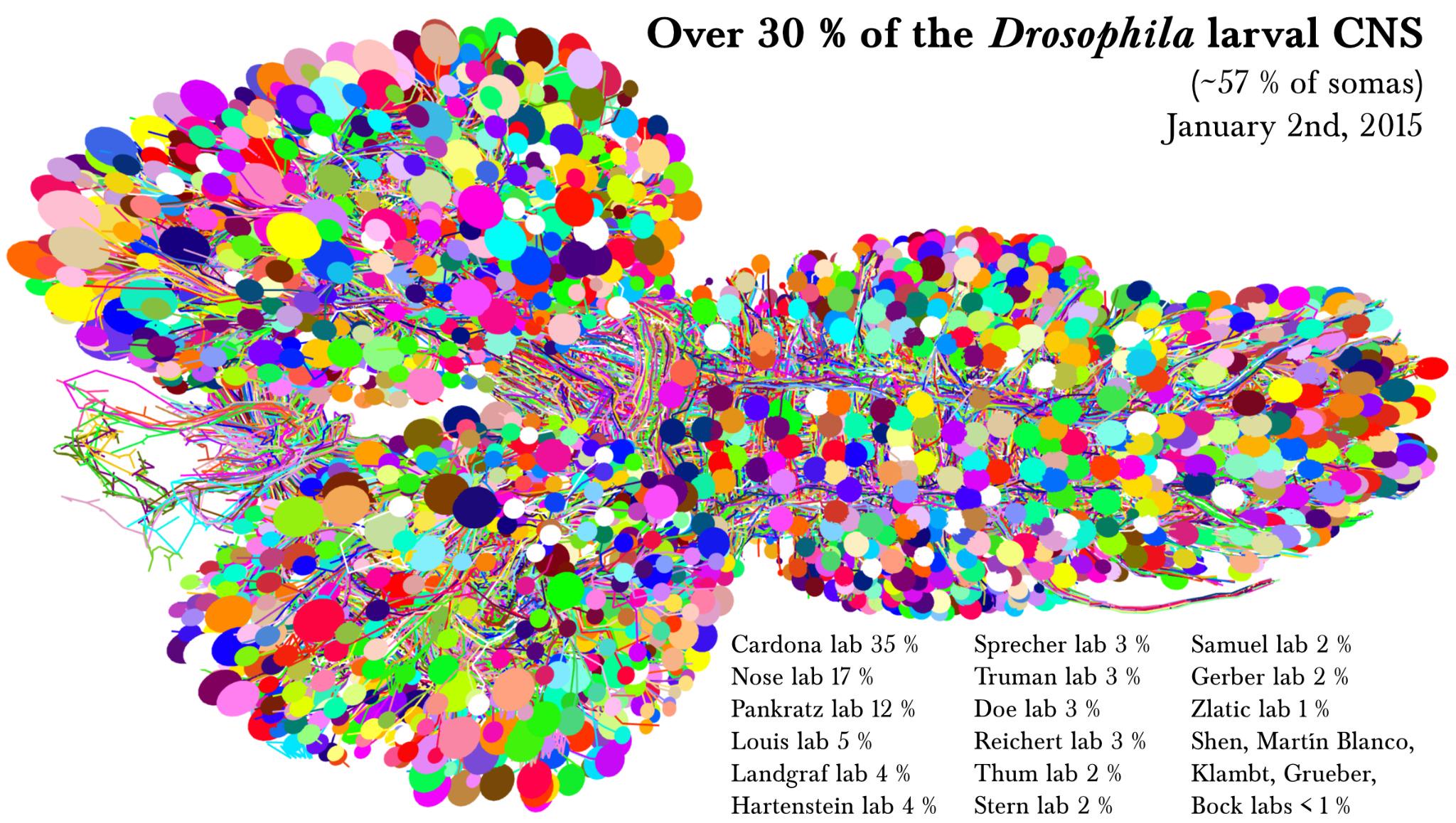
3D View of Drosophila Larval CNS Tracing by Albert Cardona’s lab at HHMI Janelia Research Campus and collaborators.¶
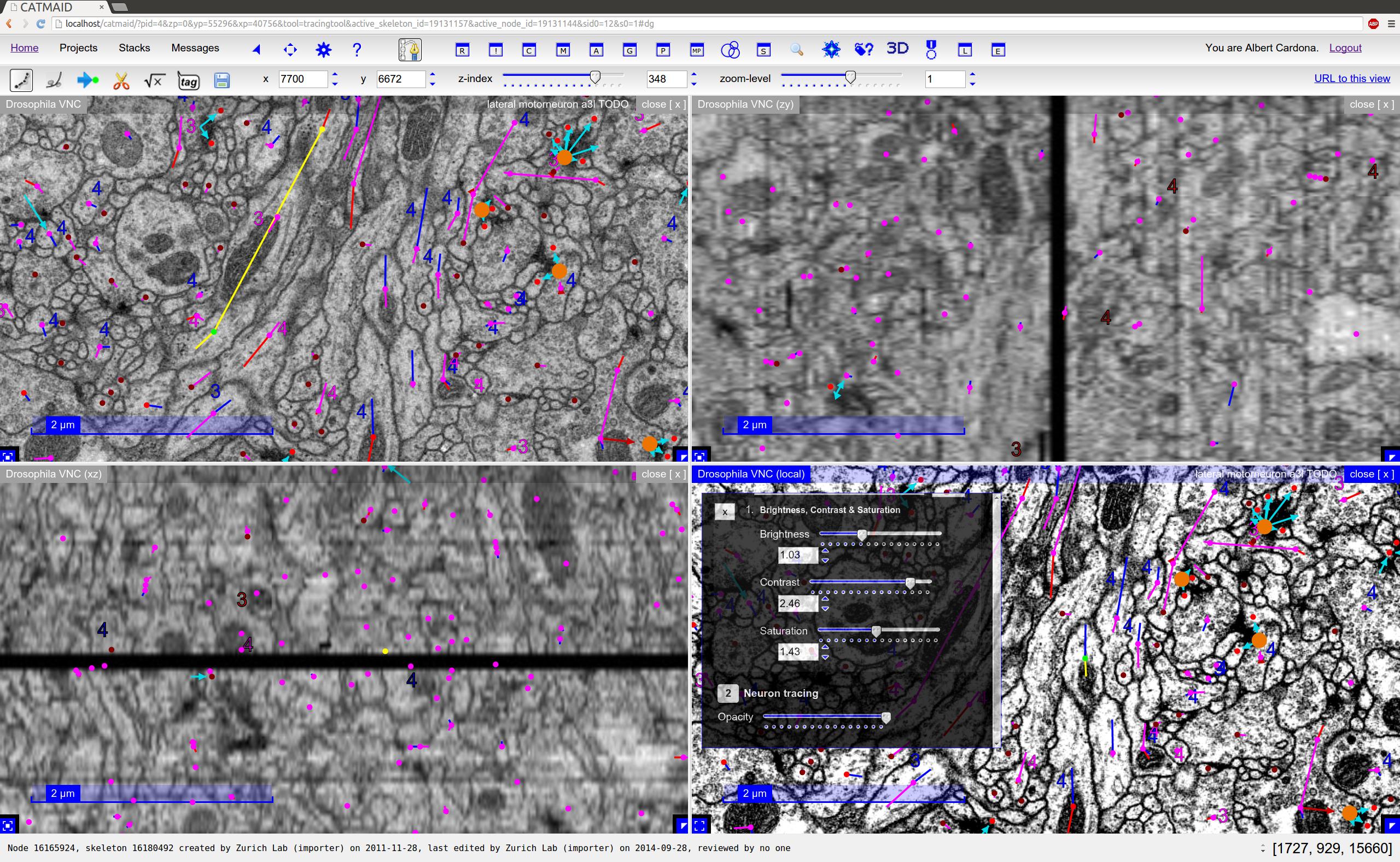
Orthogonal views of the same image stack are supported by CATMAID¶
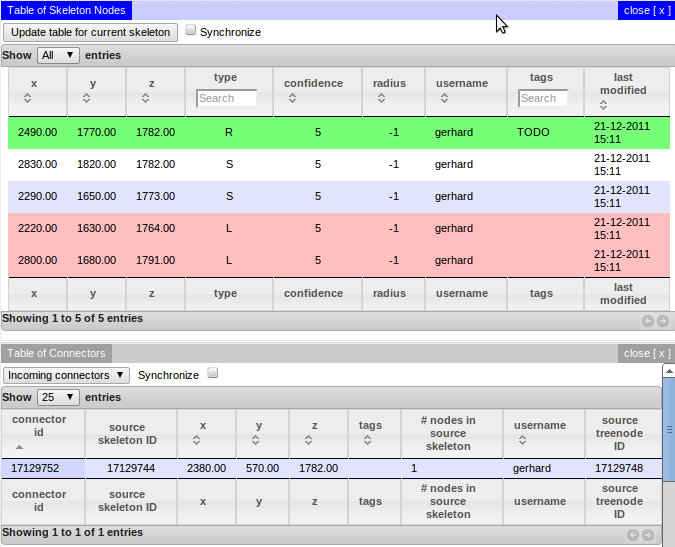
CATMAID’s Skeleton Node and Connector Table¶
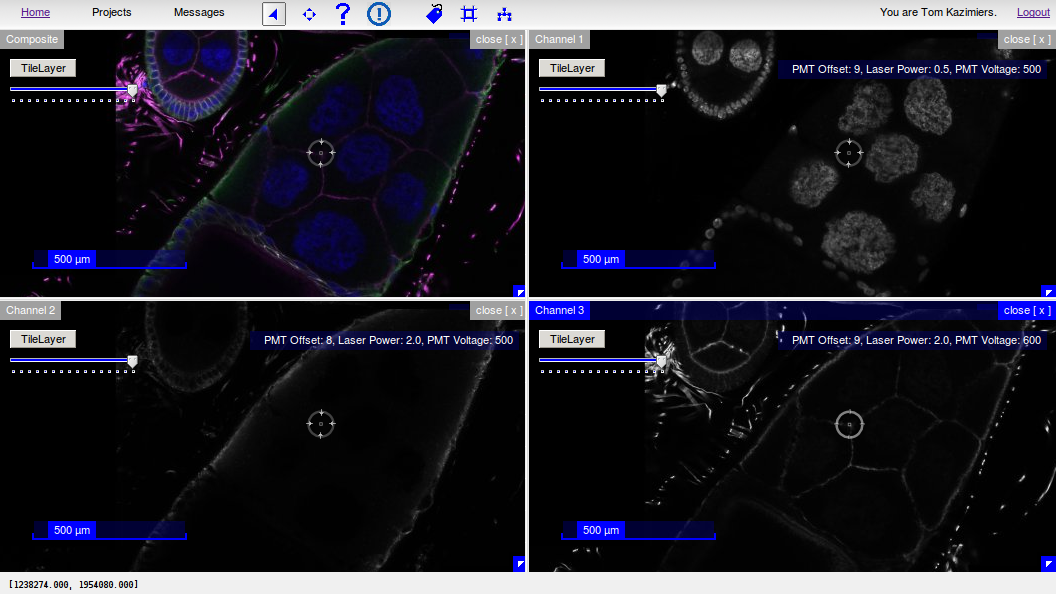
CATMAID’s Selector tool: Mirrored mouse cursor in all open stacks¶
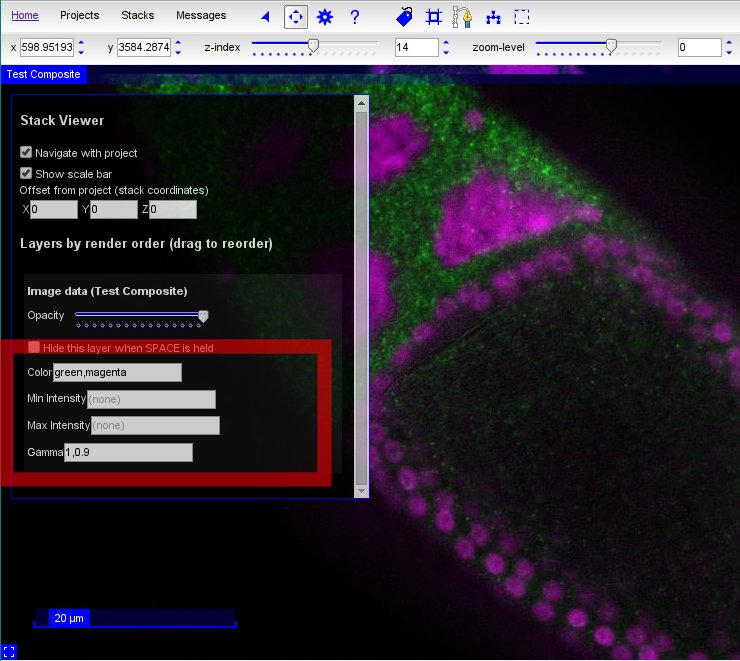
Some tile sources in CATMAID support dynamic settings

Landmark based transformation of neurons on the right side of the Drosophila larva brain to its left side and NBLAST matching for identification.¶
